Giant Virus Revealed in 3-D Using X-ray Laser
Experiment Compiles Hundreds of Images, Reveals Inner Details of Intact ‘Mimivirus’
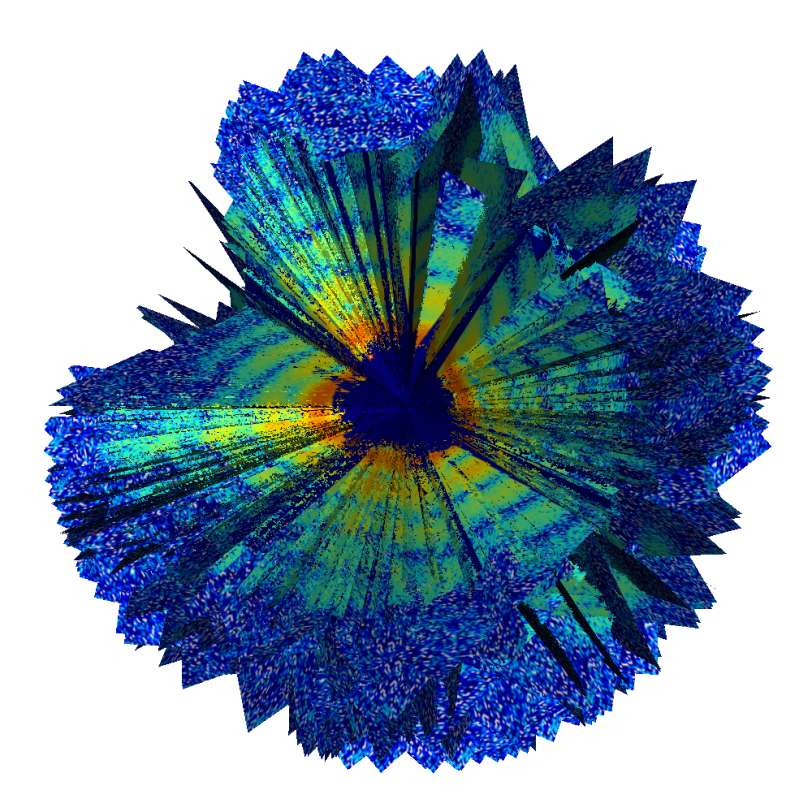
For the first time, researchers have produced a 3-D image revealing part of the inner structure of an intact, infectious virus, using a unique X-ray laser at the Department of Energy’s SLAC National Accelerator Laboratory. The virus, called Mimivirus, is in a curious class of “giant viruses” discovered just over a decade ago.
The experiment at SLAC’s Linac Coherent Light Source (LCLS), a DOE Office of Science User Facility, establishes a new technique for reconstructing the 3-D structure of many types of biological samples from a series of X-ray laser snapshots.
“Ever since I started in this field of X-ray laser research, this has always been the dream – to acquire 3-D images of real biological samples,” said Tomas Ekeberg, a biophysicist at Uppsala University in Sweden and lead author of the study, published March 2 in Physical Review Letters. “This is fantastic – it’s a breakthrough in our research.”
Mysterious World of Giant Viruses
Mimivirus is so big – its volume is thousands of times larger than the smallest viruses and even larger than some bacteria – that it was misclassified as a bacterium until 2003. Subsequent discoveries have found other giant viruses, some of which are even larger.
Mimivirus is also genetically complex, with nearly 1,000 major genes compared to only a handful in the HIV virus.
Scientists have been trying to determine the inner structure of these giant viruses to learn more about their origins: For example, did they borrow genes over time from the host organisms they infect, like amoebas? Did they precede cell-based life or devolve from cell-based organisms?
Light Pattern Portrait
In the LCLS experiment, researchers sprayed a gas-propelled aerosol containing active Mimivirus samples in a thin stream into the X-ray laser beam, which scattered off the viruses and produced light patterns on a detector that were recorded as diffraction images.
Researchers customized sophisticated analysis software developed at Cornell University to compile hundreds of individual images from separate virus particles into a single 3-D portrait showing the general shape and inner features of Mimivirus. Each image captured a projection of a separate virus particle at a random orientation, so the collection of images of viruses in different orientations provided a more complete, 3-D view.
While the technique used at LCLS did not provide high-resolution details of the internal virus structure, it did confirm that its contents are lopsided, with an area that appears more densely concentrated.
“We can see quite clearly that the inside of these viruses is not uniform,” Ekeberg said.
This same general feature had also been seen before using an electron-based imaging technique with frozen samples, and LCLS allows studies of viruses and other biological samples in a more natural, intact state. Researchers said that LCLS shows promise for achieving sharper images that reveal more inner details in the future because of the uniquely intense, penetrating power of its X-rays.
3-D Vision for X-ray Laser Studies
The same technique was recently used to study bacterial cell structures. LCLS managers have launched an initiative with the scientific community to improve techniques for imaging intact, biological particles that are difficult to study.
Janos Hajdu, a professor of biophysics at Uppsala and a pioneer in biological particle imaging with X-ray lasers, said the research team plans to apply the 3-D imaging technique to other types of samples and to improve the image quality. He said, “The next Holy Grail is to study large, single proteins at LCLS.”
Participants in the research included scientists from SLAC’s LCLS, Lawrence Berkeley National Laboratory and Kansas State University; Uppsala University in Sweden; Aix Marseille University and CEA-Saclay in France; the Center for Free-Electron Laser Science at DESY, University of Hamburg in Germany, PNSensor GmbH, Max Planck Institute’s Semiconductor Laboratory, Max Planck Institute for Extraterrestrial Physics in Germany, and the European XFEL in Germany; National University of Singapore; and The University of Melbourne in Australia. The work was supported by the Swedish Research Council, the Knut and Alice Wallenberg Foundation, the European Research Council, the Röntgen-Ångström Cluster, and Stiftelsen Olle Engkvist Byggmästare.
Citation: T. Ekeberg et al., Physical Review Letters, 2 March 2015 (10.1103/PhysRevLett.114.098102)
For questions or comments, contact the SLAC Office of Communications at communications@slac.stanford.edu.
SLAC is a multi-program laboratory exploring frontier questions in photon science, astrophysics, particle physics and accelerator research. Located in Menlo Park, Calif., SLAC is operated by Stanford University for the U.S. Department of Energy’s Office of Science.
SLAC National Accelerator Laboratory is supported by the Office of Science of the U.S. Department of Energy. The Office of Science is the single largest supporter of basic research in the physical sciences in the United States, and is working to address some of the most pressing challenges of our time. For more information, please visit science.energy.gov.